Alotaibi Lab. on Transcriptional Regulation
OVERVIEW
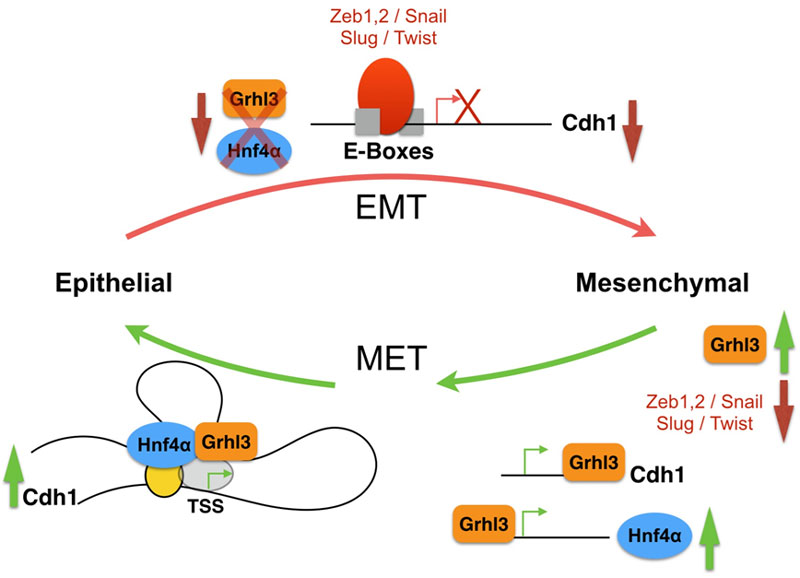
The epithelial to mesenchymal transition (EMT) and mesenchymal to epithelial transition (MET) are crucial for proper embryonic development but are also exploited by tumor cells. Tumor cells of epithelial origin can benefit from an EMT event and lose their epithelial and polarized morphology and gain mesenchymal unpolarized cell morphology. These cells can now cross the blood barrier, relocate and colonize sites in distant organs. Eventually, they will initiate MET to become epithelial-like and successfully form secondary tumors. Induction of EMT is integrated by many different signals but shares the activation of intracellular EMT-inducers like Snail, Slug, Zeb1 and others, which are transcription factors acting as repressors of the Ca2+-dependent adhesion molecule (E-cadherin), which is at the center of EMT-MET, its expression must be successfully downregulated for EMT to proceed, and must be rapidly restored for proper MET.
To date, MET is considered as a reverse process of EMT. However, MET is not simply the reverse process of EMT. It is a complicated and dynamic process. There must be a more complex regulatory mechanisms ensuring the establishment of an epithelial phenotype. This dynamic MET process can only in part be explained by the upregulation and downregulation of EMT inducers. It suggests the presence of other transcriptional activators for E-cadherin, which could also be regulators of MET.
RESEARCH INTERESTS
We are interested in understanding the genetic and molecular mechanisms of tumorigenesis, with a focus on dissecting the transcriptional networks controlling the mesenchymal to epithelial transition.
Transcriptional networks have been described before; examples include, but are not limited to, the transcriptional network of EMT, the core pluripotency network of embryonic stem cells, and the transcriptional network of blood stem cells. There are different kinds (or motifs) of transcriptional regulatory networks. These are sets of transcription factors that control the initiation and maintenance of certain transcriptional loops or chains to augment or stabilize gene expression. Examples of the network motifs include for example autoregulatory loops, where a transcription factor would be regulating its own expression, such as the Nanog loop in the core pluripotency network. In feed forward loops, transcription factor X will regulate both Gene Y and Gene Z, where Gene Y encodes a transcription factor that also regulates Gene-Z, an example of such regulatory loop comes from our recent study describing the initiation of MET, in which Grhl3 regulates the expression of both Cdh1 and Hnf4a, Hnf4a is then recruited to the Cdh1 chromatin to boost up the Grhl3-dependent regulation. In a multi-component loop, transcription factors regulate the expression of their regulators in a positive feedback loop; a very well-known example is the closed circuit of Oct4/Sox2 in embryonic stem cells regulatory network. In a regulatory chain, three or more transcription factors are ordered in a series, the chain ends when the transcription factor has no target (i.e. no transcription factor target) or is autoregulatory, an example of such regulatory chains comes from the hematopoietic stem cells, where b-catenin regulates HoxB4, which in turn activates the expression of c-myc. Common to most transcriptional network studies is the methodology used; most share similar sets of experimental and knowledge-based approaches, combining data mining, gene expression profiling and gene silencing, all integrated into a powerful computational analysis to correctly decipher transcriptional nodes and network motifs.
In order to validate the existence of an MET network, we will investigate in detail the expression profiles of several EMT-MET cellular models; including NMuMG cells as well as human cancer cell line models that are known to undergo mesenchymal to epithelial transition. We will use an integrated approach, including data mining, high throughput expression profiling, gene silencing, and validation by protein-protein and protein-DNA interaction techniques. As a result, we anticipate defining an epithelial state specific transcription factor gene set. Using this gene set we will continue to define the relationships between the members and their function within a network. Our preliminary data suggest the existence of several overlapping transcriptional nodes having Grhl3 in the center of a larger network and E-cadherin as the major outcome. Our main goal is to confirm the transcriptional relationships between the identified transcription factors and to draw a more comprehensive picture of the MET network. As a result, we aim to identify a core MET network, which, in the long run, may lead to the identification of therapeutic targets for the control of MET during metastasis. MET is also a critical step during the reprogramming of iPSCs, thus identifying a core MET network could be utilized to improve the efficiency and quality of the reprogramming process.
RESEARCH HIGHLIGHTS
To date, there is a substantial amount of data in the literature analyzing the EMT program both during embryonic development and also during tumorigenesis, but a similar understanding of the MET program is still lagging. Recently we have identified a feed forward loop, which is composed of the transcription factors Grhl3 and Hnf4a that is essential for the progression of MET. In fact, Grhl3 emerged as an absolute requirement for the initiation of MET. Cells lacking Grhl3 as a result of RNAi-mediated knockdown failed to initiate the MET program. This is in part due to their inability to upregulate E-cad expression. Furthermore, we found that Grhl3 contributes to the regulation of Hnf4α by binding to a well-conserved motif present within the Hnf4α promoter P2. We believe that there is a need to gather more credible scientific evidence that answers key questions regarding how the process of MET is regulated, how it is initiated and the transcription factors involved in this regulation.
Group Members

Alotaibi Lab. on Transcriptional Regulation
Research Group Leader
Hani ALOTAİBİ
hani.alotaibi@ibg.edu.tr
+90 232 299 41 00
(5071)
+9
0 232 299 41 57

Canan Büşra ERSAYAR
Research Technician
canan.ersayar@ibg.edu.tr
0 (232) 299 _ _

İNCİ YAPRAK
MSc Student
inci.yaprak@ibg.edu.tr

Gkamze IMPRAIMOGLOU KAFAZ
MSc Student
gkamze.impraimoglou@ibg.edu.tr

Zehra Yağmur ALAGÖZ
Undergraduate Student
zehra.alagoz@ibg.edu.tr

Esin ŞEKEROĞLU
PhD Student
esin.ozkuru@ibg.edu.tr

Seray YETKİN
PhD Student
seray.yetkin@ibg.edu.tr

ŞEYMA NUR YILDIZ
PhD Student
seyma.yildiz@ibg.edu.tr

Zeynep KARAGÖZ
MSc Student
zeynep.karagoz@ibg.edu.tr

Hilal ÇAĞLAYAN
PhD Student
hilal.caglayan@std.ibg.edu.tr

Helin SAĞIR
PhD Student
helin.sagir@ibg.edu.tr

İNCİ YAPRAK
PhD Student
inci.yaprak@ibg.edu.tr

İNCİ YAPRAK
PhD Student
inci.yaprak@ibg.edu.tr

İNCİ YAPRAK
PhD Student
inci.yaprak@ibg.edu.tr

Esra DİNCASLAN
MSc Student
esra.dincaslan@std.ibg.edu.tr
Former Members

Burcu ŞENGEZ
Research Assistant
burcu.sengez@ibg.edu.tr

Tuğşen AYDEMİR
Post-Doc Researcher
tugsen.aydemir@msfr.ibg.edu.tr

Çağrı BULDU
MSc Student
cagri.buldu@msfr.ibg.edu.tr

Diana ZEID
MSc Student
diana.zeid@msfr.ibg.edu.tr

Hadi ROUHRAZİ
Post-Doc Researcher
hadi.rouhrazi@ibg.edu.tr

Naz ŞERİFOĞLU
MSc Student
naz.serifoglu@msfr.ibg.edu.tr

Caner YARIMÇAM
PhD Student
caner.yarimcam@msfr.ibg.edu.tr

Gürkan AKYILDIZ
Post-Doc Researcher
gurkan.akyildiz@msfr.ibg.edu.tr

Hazal AYTEKİN
PhD Student
hazal.aytekin@msfr.ibg.edu.tr

İrem KÜLLÜ
Undergraduate Student
irem.kullu@msfr.ibg.edu.tr

Tuğçe YAPAR
PhD Student
tugce.yapar@ibg.edu.tr

Ayça ÖZKAN
MSc Student
ayca.ozkan@ibg.edu.tr
0 (232) 299 _ _

Şimal KAYIKÇI
Undergraduate Student
simal.kayikci@ibg.edu.tr

Helin SAĞIR
PhD Student
helin.sagir@ibg.edu.tr

Seray YETKİN
MSc Student
seray.yetkin@ibg.edu.tr

Sevgi ERDEM
Visiting Researcher
sevgi.erdem@ibg.edu.tr

İLAYDA KAYA
Undergraduate Student
ilayda.kaya@msfr.ibg.edu.tr

Filiz ŞAHİN
MSc Student
filiz.sahin@ibg.edu.tr

Doğa ESKİER
PhD Student
doga.eskier@ibg.edu.tr

Şehriban BÜYÜKKILIÇ
PhD Student
sehriban.buyukkilic@ibg.edu.tr

Nilüfer DÜZ
PhD Student
nilufer.duz@ibg.edu.tr

Cemre Su YENİCELİ
Undergraduate Student
cemresu.yeniceli@ibg.edu.tr

Elif Bilge KİRİŞ
Undergraduate Student
elif.kiris@std.ibg.edu.tr
Selected Publications
Doğa Eskier, Seray Yetkin, Nazmiye Arslan, Gökhan Karakülah, Hani Alotaibi. Exploring Regulatory Roles of Transposable Elements in EMT and MET through Data-Driven Analysis: Insights from regulaTER. Journal of Molecular Biology. 2025 January ; 437 (2) : 168887. doi:10.1016/j.jmb.2024.168887. Download
Abu Alhaija AA, Lone IN, Ozkuru Sekeroglu E, Batur T, Angelov D, Dimitrov S, Hamiche A, Firat Karalar EN, Ercan ME, Yagci T, Alotaibi H, Diril MK. Development of a mouse embryonic stem cell model for investigating the functions of the linker histone H1-4.. FEBS open bio. 2024 February . doi:10.1002/2211-5463.13750. Download
Alotaibi H, Meuwissen R, Sayan AE. Editorial: Understanding the mesenchymal to epithelial transition: a much needed angle for epithelial mesenchymal plasticity.. Frontiers in cell and developmental biology. 2024 October ; 12 : 1497515. doi:10.3389/fcell.2024.1497515. Download
Alotaibi H. The transcription factor ELF5 is essential for early preimplantation development.. Molecular biology reports. 2023 March ; 50 : 2119-2125. doi:10.1007/s11033-022-08217-z. Download
Yetkin S, Alotaibi H. Selection and validation of novel stable reference genes for qPCR analysis in EMT and MET.. Experimental cell research. 2023 July ; 428 (1) : 113619. doi:10.1016/j.yexcr.2023.113619. Download
Demir AB, Baris E, Kaner UB, Alotaibi H, Atabey N, Koc A. Toll-interacting protein may affect doxorubicin resistance in hepatocellular carcinoma cell lines.. Molecular biology reports. 2023 August . doi:10.1007/s11033-023-08737-2. Download
Sengez B, Carr BI, Alotaibi H. EMT and Inflammation: Crossroads in HCC.. Journal of gastrointestinal cancer. 2022 January . doi:10.1007/s12029-021-00801-z. Download
Toyran N. Alotaibi H.. Evaluating The Transcriptional Regulation of Cdh1 by Grhl3 in Different Cellular Models. Journal of The Institute of Science and Technology. 2022 March ; 12 (1) : 446-454. doi:10.21597/jist.963916. Download
Hani Alotaibi. Crif1 is Required for Proper Mesenchymal to Epithelial Transition. Cumhuriyet Science Journal. 2022 January ; 43 (2) : 165. doi:10.17776/csj.1062126. Download
Evin Iscan, Umut Ekin, Gokhan Yildiz, Ozden Oz, Umur Keles, Aslı Suner, Gulcin Cakan-Akdogan, Gunes Ozhan, Marta Nekulova, Borivoj Vojtesek, Hamdiye Uzuner, Gökhan Karakülah, Hani Alotaibi and Mehmet Ozturk. TAp73β Can Promote Hepatocellular Carcinoma Dedifferentiation. Cancers. 2021 February ; 13 (4) : 783. doi:10.3390/cancers13040783. Download
Toy HI, Karakülah G, Kontou PI, Alotaibi H, Georgakilas AG, Pavlopoulou A. Investigating Molecular Determinants of Cancer Cell Resistance to Ionizing Radiation Through an Integrative Bioinformatics Approach.. Frontiers in cell and developmental biology. 2021 April ; 9 : 770. doi:10.3389/fcell.2021.620248. Download
Tugce Batur, Ayse Argundogan, Umur Keles, Zeynep Mutlu, Hani Alotaibi, Serif Senturk, Mehmet Ozturk. AXL knock-out in SNU475 hepatocellular carcinoma cells provides evidence for lethal effect associated with G2 arrest and polyploidization. International Journal of Molecular Sciences. 2021 December ; 22 (24) : 13247. doi:10.3390/ijms222413247. Download
Hani Alotaibi. Transcriptional impact of E-cadherin loss on embryonic stem cells. Jordan Journal of Biological Sciences. 2021 December ; 14 (5) . doi:10.54319/jjbs/140524. Download
Ekin U, Yuzugullu H, Ozen C, Korhan P, Bagirsakci E, Yilmaz F, Yuzugullu OG, Uzuner H, Alotaibi H, Kirmizibayrak PB, Atabey N, Karakülah G, Ozturk M. Evaluation of ATAD2 as a Potential Target in Hepatocellular Carcinoma.. Journal of gastrointestinal cancer. 2021 November . doi:10.1007/s12029-021-00732-9. Download
Lone IN, Sengez B, Hamiche A, Dimitrov S, Alotaibi H. The Role of Histone Variants in the Epithelial-To-Mesenchymal Transition.. Cells. 2020 November ; 9 (11) . doi:10.3390/cells9112499. Download
Sengez B, Aygün I, Shehwana H, Toyran N, Tercan Avci S, Konu O, Stemmler MP, Alotaibi H. The Transcription Factor Elf3 Is Essential for a Successful Mesenchymal to Epithelial Transition.. Cells. 2019 August ; 8 (8) . doi:10.3390/cells8080858. Download
Alotaibi H, Tuzlakoğlu-Öztürk M, Tazebay UH. The Thyroid Na+/I- Symporter: Molecular Characterization and Genomic Regulation. MOLECULAR IMAGING AND RADIONUCLIDE THERAPY. 2017 February ; 26 (1) : 92-101. doi:10.4274/2017.26.suppl.11. Download
İmge KUNTER, Emine KANDEMİŞ, Hani ALOTAİBİ, Tülay CANDA, Esra ERDAL BAĞRIYANIK. Alteration in the subcellular location of the inhibitor of growth protein p33(ING1b) in estrogen receptor alpha positive breast carcinoma cells. Turkish Journal of Biology. 2017 ; 41 (1) : 105-112. doi:10.3906/biy-1602-95 . Download
Alotaibi H, Basilicata MF, Shehwana H, Kosowan T, Schreck I, Braeutigam C, Konu O, Brabletz T, Stemmler MP. Enhancer cooperativity as a novel mechanism underlying the transcriptional regulation of E-cadherin during mesenchymal to epithelial transition. Biochimica et biophysica acta. 2015 June ; 1849 (6) : 731-42. doi:10.1016/j.bbagrm.2015.01.005. Download
Bedzhov I, Alotaibi H, Basilicata MF, Ahlborn K, Liszewska E, Brabletz T, Stemmler MP. Adhesion, but not a specific cadherin code, is indispensable for ES cell and induced pluripotency.. Stem cell research. 2013 November ; 11 (3) : 1250-63. doi:10.1016/j.scr.2013.08.009. Download
Telkoparan P, Erkek S, Yaman E, Alotaibi H, Bayık D, Tazebay UH. Coiled-coil domain containing protein 124 is a novel centrosome and midbody protein that interacts with the Ras-guanine nucleotide exchange factor 1B and is involved in cytokinesis.. PloS one. 2013 July ; 8 (7) : e69289. doi:10.1371/journal.pone.0069289. Download
Mumcuoglu M, Bagislar S, Yuzugullu H, Alotaibi H, Senturk S, Telkoparan P, Gur-Dedeoglu B, Cingoz B, Bozkurt B, Tazebay UH, Yulug IG, Akcali KC, Ozturk M. The ability to generate senescent progeny as a mechanism underlying breast cancer cell heterogeneity.. PloS one. 2010 June ; 5 (6) : e11288. doi:10.1371/journal.pone.0011288. Download
Alotaibi H, Yaman E, Salvatore D, Di Dato V, Telkoparan P, Di Lauro R, Tazebay UH. Intronic elements in the Na+/I- symporter gene (NIS) interact with retinoic acid receptors and mediate initiation of transcription.. Nucleic acids research. 2010 June ; 38 (10) : 3172-85. doi:10.1093/nar/gkq023. Download
Alotaibi H, Ricciardone MD, Ozturk M. Homozygosity at variant MLH1 can lead to secondary mutation in NF1, neurofibromatosis type I and early onset leukemia.. Mutation research. 2008 January ; 637 (1-2) : 209-14. doi:10.1016/j.mrfmmm.2007.08.003. Download
Alotaibi H, Yaman EC, Demirpençe E, Tazebay UH. Unliganded estrogen receptor-alpha activates transcription of the mammary gland Na+/I- symporter gene.. Biochemical and biophysical research communications. 2006 July ; 345 (4) : 1487-96. doi:10.1016/j.bbrc.2006.05.049. Download
Total : 25
Selected Book Chapters
TRANSCRIPTIONAL REGULATION OF EPITHELIAL-TO-MESENCHYMAL TRANSITION BY TGFb ISOFORMS (2021). RESEARCH & REVIEWS IN HEALTH SCIENCES. Gece Kitaplığı.
Kodom Optimizasyonu Ve Rekombinant Protein Üretimindeki Yeri (2021). PROTEİN: YAPISI, MÜHENDİSLİĞİ, ETKİLEŞİMLERİ, DİNAMİĞİ VE İLAÇ TASARIMINDAKİ YERİ. Ankara Nobel Tıp Yayınevi.
Molecular Mechanisms of Hepatocellular Carcinoma (2016). Hepatocellular Carcinoma: Diagnosis and Treatment, 3rd Edition. Springer.
Total : 3
Projects
The Scientific and Technological Research Council of Turkey - TUBITAK - RD : Kanser ve Osteoporoz Tedavisi İçin Monoklonal Antikor Etkin Maddeli Biyobenzer İlaç Geliştirilmesi ve Üretilmesi, Ongoing
The Scientific and Technological Research Council of Turkey - TUBITAK - RD : Mezenkimal-Epitelyal Dönüşümünü (Met) Kontrol Eden Transkripsiyonel Ağların Belirlenmesi, Finished
DEÜ BAP - Dokuz Eylul University Scientific Research Projects Coordination Unit - RD : Mezenkimal-Epiteliyal Sürecinde ELF3'ün GRHL3 Ekspresyonuna Olan Etkisinin Belirlenmesi, Finished
The Scientific and Technological Research Council of Turkey - TUBITAK - RD : Identification of Transcription Factors Influencing the Epigenetic Reprogramming During the Mesenchymal to Epithelial Transition (MET), Finished
The Scientific and Technological Research Council of Turkey - TUBITAK - RD : Epigenetics of Epithelial-to-Mesenchymal and Mesenchymal-to-Epithelial Transitions : Role of Histone Variants and Pioneer Transcription Factors, Ongoing
The Scientific and Technological Research Council of Turkey - TUBITAK - RD : Functional Analysis of Long Non-coding RNAs Critical for the Mesenchymal to Epithelial Transition, Ongoing
The Scientific and Technological Research Council of Turkey - TUBITAK - RD : HepatoselülerKarsinoma Hücre Hatlarının Doksorubisin Hassasiyetini Arttırmaya Yönelik TOLLIP Geninin Doksorubisin Dirençliliğindeki Rolünün Belirlenmesi, Ongoing
The Scientific and Technological Research Council of Turkey - TUBITAK - RD : Epigenetics of the Mesenchymal to Epithelial Transition, Ongoing
The Scientific and Technological Research Council of Turkey - TUBITAK - RD : Development of a Computational Tool to Predict Transposable Elements With Gene Regulatory Roles and Identification of Transposable Elements That Are Involved in to Mesenchymal – Epithelial Transition, Ongoing
The Scientific and Technological Research Council of Turkey - TUBITAK - RD : Mezenkimal Epitelyal Dönüşüm Sürecinde Rol Alan mikroRNA'ların Tanımlanması ve Fonksiyonel Analizleri, Ongoing
Health Institutes of Turkey- TUSEB - RD : Meme Kanseri MCF-7 Hücre modelinde ZEB1 ile İndüklenen EMT/MET Süreçlerindeki Kritik LncRNA’ların Tanımlanması ve Fonksiyonel Analizleri, Ongoing
The Scientific and Technological Research Council of Turkey - TUBITAK - RD : Sağlıkta ve Rahman Sendromunda Bağlayıcı Histon H1E, Ongoing
The Scientific and Technological Research Council of Turkey - TUBITAK - RD : Desminin kromatine ile etkileşiminin araştırılması, Finished
DEÜ BAP - Dokuz Eylul University Scientific Research Projects Coordination Unit - RD : Kanser ve metastaz oluşumunda Elf5 genin rolünün, E-cadherin üzerindeki regülasyonu ve EMT/MET kontrol mekanizmasının araştırılması., Ongoing
The Scientific and Technological Research Council of Turkey - TUBITAK - RD : Determination of the role of TP73 in AXL regulation and examining the TP73-AXL relationship in DNA damage, Ongoing
The Scientific and Technological Research Council of Turkey - TUBITAK - RD : KORUNMA VE TEDAVİ ULUSAL PLATFORMU, Ongoing
The Scientific and Technological Research Council of Turkey - TUBITAK - RD : The Role of the C730027H18Rik Gene in Mesenchymal-Epithelial Transition: A Novel Perspective on MET Regulation, Ongoing
Awards
- Project Performance Award by TUBITAK, 2021
- Short Term Fellowship by EMBO, 2005
Academic Memberships
- Biochemical Society, 2000
Contact

Alotaibi Lab. on Transcriptional Regulation
Research Group Leader
Hani ALOTAİBİ
hani.alotaibi@ibg.edu.tr
+90 232 299 41 00
(5071)
+9
0 232 299 41 57
