Dimitrov Lab. on Chromatin Biology and Epigenetics
OVERVIEW
We are working in the field of chromatin epigenetics, one of the most exciting field of modern biology. Our interests are focused in unveiling the epigenetic mechanisms used by the cells under both normal and disease conditions. This is achieved through the use of an original and largely non-conventional combination of genome-wide methods, a plethora of physicochemical, biochemistry, molecular and cell biology techniques, mouse genetics and high resolution structural approaches, including X-ray diffraction and electron cryo-microscopy.
SPECIFIC GOALS
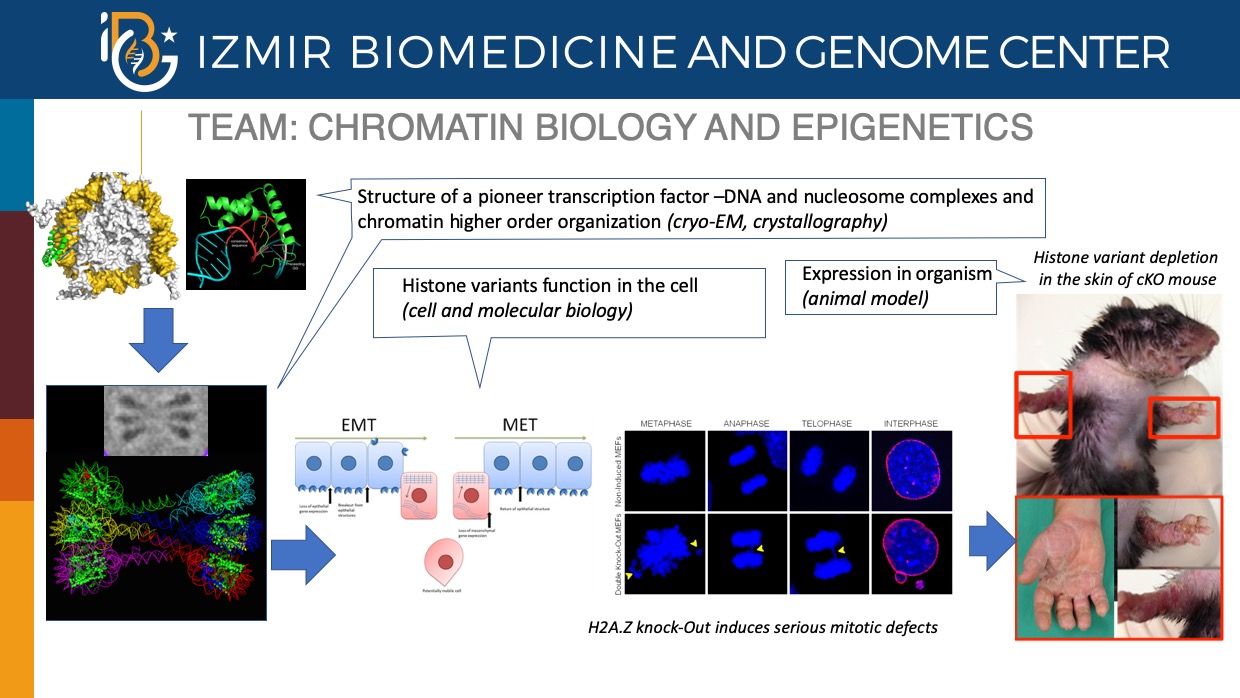
The condensed chromatin fibers and in particular, the nucleosome, represent a barrier for the proteins to bind to their cognate DNA sequence and thus, to function. The cell has elaborated very sophisticated epigenetic tools and strategies to overcome the chromatin barrier and to “extract” the necessary information from specific chromatin loci. Among them, histone variants, the non-allelic isoforms of conventional histones, are key players. Incorporation of histone variants confers novel structural and functional properties of the nucleosome, but how this is achieved and altered in diseases remains obscure. Our main interest is to understand the function of histone variants in both normal and disease conditions.
Our current projects are centered on:
- Analysis of the in vivo roles of histone variants in the formation and maintenance of specific histone variant-mediated chromatin structures and how this structures contribute to cell and organ homeostasis.
- Implication of histone variants in Epithelial-to-Mesenchymal (EMT) and Mesenchymal-to-Epithelial Transitions (MET), essential processes in both mammalian development and cancer progression.
- Dissection of the specific mechanisms used by primary transcription factors (PTFs) , able to invade highly compact chromatin, to remodel it and consequently, to allow transcription of specific EMT or MET genes to proceed.
- Mechanisms of assembly of condensed chromatin fiber and impact of histone variants
Selected publications (2010-2019)
- Garcia-Saez et al. (2018) Molecular Cell. 2018 Dec 6;72(5):902-915.e7. doi: 10.1016/j.molcel.2018.09.027.
- Bednar, J. et al. (2017) «Structure and dynamics of the nucleosome bound to linker histone H1» Mol. Cell 66, 384-397 (ranked by Faculty 1000, “F1000 Prime”)
- Papin, A.et al. «Combinatorial DNA methylation code at repetitive elements» Genome Research, 2017 Mar 27. pii: gr.213983.116. doi: 10.1101/gr.213983.116. [Epub ahead of print]
- Roulland, Y., et al. «The flexible ends of CENP-A nucleosome are required for mitotic fidelity», Mol. Cell, 2016, 18;63(4):674-85.
- Latryck, C., et al. «YL1, H2A.Z-specific histone chaperone, required for ATP-dependent deposition of H2A.Z in chromatin», Nature Struct & Mol. Biol. 2016 Apr;23(4):309-16.
- Obri, A., et al. (2014) «ANP32E is a histone chaperone that removes H2A.Z from chromatin» Nature (full article), 505(7485):648-53.
- Goutte-Gattat, D. et al. (2013) «The phosphorylation of the CENP-A NH2-terminus in mitotic centromeric chromatin is required for kinetochore function», Procl .Natl. Acad. USA110, 8579-8584
- Mayer, S et al. (2011) «From crystals, footprints and cryo-electromicrographs to large and soft structures: nanoscale modeling of the nucleosomal stem» Nucleic Acids Research 39(21):9139-54
- Montel, F., et al. (2011) «RSC remodelling of oligonucleosomes: an atomic force microscopy study», Nucl. Acids. Res. 39(7), 2571-2570
- Chantalat, S. et al. (2011) «Histone H3 at lysine 36 is involved in the formation of constitutive and facultative heterochormatin», Genome Research 21(9):1426-37
- Syed, S. H., et al. (2010) «Single base resolution mapping of H1-nucleosome interactions and 3D organization of the nucleosome», Proc. Natl. Acad. Sci. USA107, 9620-9625
- Shuaib, M. et al. (2010) «HJURP binds CENP-A via a highly conserved N-terminal domain and mediates its deposition to centromeres» Proc.Natl. Acad. Sci.USA 107(4):1349-54
- Shukla, M., (2010) «Remosomes: novel RSC generated non-mobilized particles with ~180 bp of DNA loosely associated with the histone octamer», Proc. Natl. Acad. Sci. USA107(5):1936-41 (ranked with score 12 at Faculty 1000)
Group Members

Dimitrov Lab. on Chromatin Biology and Epigenetics
Former Members

Stefan DIMITROV
Research Group Leader
stefan.dimitrov@ibg.edu.tr

Esra TÜRKER
Researcher
esra.turker@ibg.edu.tr

Imtiaz Nisar LONE
Researcher
imtiaznisar.lone@ibg.edu.tr

Ceren ŞAHİN
Technician
ceren.sahin@ibg.edu.tr

Esin ŞEKEROĞLU
PhD Student
esin.ozkuru@ibg.edu.tr

TUĞBA ŞEHİTOĞULLARI
MSc Student
tugba.sehitogullari@ibg.edu.tr

Burcu ŞENGEZ
Post-Doc Researcher
burcu.sengez@ibg.edu.tr

Tuğçe BATUR
Researcher
tugce.batur@ibg.edu.tr

Stefan DIMITROV
Visiting Researcher
stefan.dimitrov@ibg.edu.tr

ŞEYMA NUR YILDIZ
PhD Student
seyma.yildiz@ibg.edu.tr
Open Positions
Epigenetic Lab is seeking for Ph.D. Students
Dimitrov Lab
We are working in the field of epigenetics, one of the most exciting field of modern biology. Our interests are focused in unveiling the epigenetic mechanisms used by the cells under both normal and disease conditions. This is achieved through the use of an original and largely non-conventional combination of genome-wide methods, a plethora of physicochemical, biochemistry, molecular and cell biology techniques, mouse genetics and high resolution structural approaches.
Several Ph.D. student positions are open within the frame of the recently awarded "2232 International Grant of Outstanding Researchers" to Prof. Stefan Dimitrov. PhD student training opportunities in cutting-edge technologies will be carried out in collaboration with the labs located in various EU countries, including France. The monthly fellowship is 4500TL.
For more information on our work, please check the following link: https://scholar.google.fr/citations?user=ExgGAYoAAAAJ&hl=en
Requirements
Interested candidates holding an MSc degree should apply to IBG's PhD program (between 23-27 December, www.ibg.deu.edu.tr. We are also open to all applications from students who are already enrolled in a PhD program. For more please contact stefan.dimitrov@ibg.edu.tr
Academic Memberships
- Bulgarian Academy of Sciences, 2008
- National Academy of Sciences, India, 2016
Contact

Dimitrov Lab. on Chromatin Biology and Epigenetics