Oktay Lab. on Neuro-Genomics
Overview
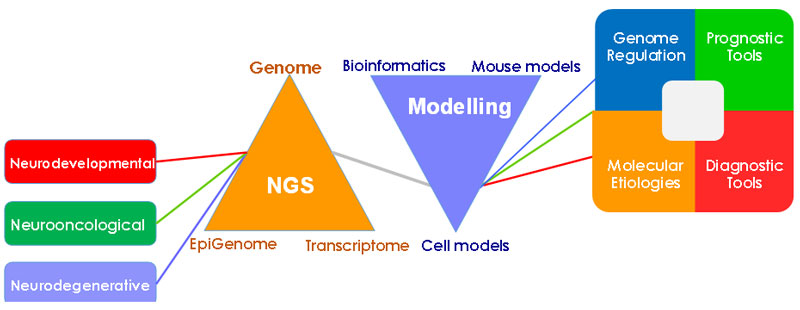
Next generation sequencing (NGS) technologies have revolutionized our approach to studying the genetic basis of diseases and caused a paradigm shift in biological sciences. With the cost of whole genome sequencing (WGS) well-below the much-anticipated $1000 level, it is now affordable to sequence the whole genome of hundreds to thousands of patients or healthy people. As a result, our knowledge of genome and its regulation in health and disease has increased tremendously: Since the completion of the Human Genome Project (HGP), millions of variants and mutations have been identified and thousands of them have been associated with both inherited and sporadic diseases. So far, an overwhelming majority of clinical genomics studies have focused on the “exome” – protein-coding parts of the genome, mainly due to its much lower cost compared to WGS. ENCODE (Encyclopedia of DNA Elements) Project and similar efforts have provided peek into the functional organization of the genome, however, our knowledge of how genomic information is utilized is far from complete and limited. A better understanding of such fundamental principles of genome regulation will undoubtedly help us explain the molecular basis of pathologic disease states. The overarching goal of our research efforts is to unravel these principles within the context of nervous system disorders.
Research Interests
The overarching goal of the Neuro-genomics Lab at IBG is the elucidation of mechanisms leading from genomic variation to diseases of the human nervous system. Our efforts can be grouped into two stages: Identification of such variants (i.e. in neurodevelopmental disorders) and their modeling in cellular and animal models with the help of genome editing tools.
Elucidation of the cellular mechanisms underlying the genetic predisposition to complex diseases and phenotypes of the nervous system is a major focus of our lab. In this respect, we utilize tools of the functional genomics to understand the molecular basis of genetic susceptibility to lethal brain tumors, namely gliomas. A multidisciplinary approach using genomic, epigenomic, transcriptomic, bioinformatic and cell biology methods is employed to achieve an integrative understanding of the early stages of gliomagenesis, as well as to identify prognostic biomarkers and therapeutic targets. We have recently identified the molecular basis of genetic predisposition to IDH-mutant gliomas and established the proto-oncogene MYC as the culprit of 8q24-associated glioma risk.
Another major focus of our research is to develop models and methods that will facilitate identification of DNA variants that cause or contribute to the etiopathogenesis of neurodevelopmental diseases. To this end, we perform whole-exome sequencing of trios and multiplex families to identify candidate variants, and test them with functional studies in cell and animal models, frequently employing CRISPR/Cas9 genome-editing tools. We have recently identified several new disease genes and variants in a large consanguineous cohort of neurodevelopmental disorders. Our current efforts are aimed towards finding therapeutic targets by utilizing a multi-omic analyses of this cohort.
A better grasp of the genotype-phenotype relationships in neurodevelopmental, as well as neurooncological, diseases is a must to translate the power of genomics into clinical benefit. Our multi-disciplinary approach will be expected to provide valuable guidance to development of therapeutic, prognostic and diagnostic tools.
Methodological Approach
- Determination of variants and mutations that cause diseases: targeted sequence analysis of genes that encode proteins and functional genome elements, RNA-sequencing, analysis of patient samples with whole exome and whole genome sequencing.
- Functional studies of identified variants and mutations in cell culture and animal models: The development of cell and animal models by CRISPR-Cas9 genome-forming technology is applied in our laboratory.
Research Highlights
Our recent work on low-grade gliomas (LGGs) has focused on understanding the genetic basis of genetic predisposition to IDH-mutated gliomas, which comprise 70-80 % of LGGs. These studies were conducted by a multi-disciplinary approach that employed transcriptomic, proteomic, immunohistochemical, clinical and epidemiologic analyses of tumor and blood samples from LGG patients. A manuscript based on these studies is under review. Also, a TUBITAK 1001 grant has recently (2015) been awarded for 3 years to study the mechanistic basis of 8q24.21-associated genetic predisposition to glioma. Within this project, neural stem cell (NSC) lines are being edited by using the CRISPR-Cas9 system to obtain isogenic cell lines that differ at only the disease-associated variants/loci, in the absence and presence of IDH1-R132H mutation due to the strict association of these variants with IDH1/2 mutations. An integrated analysis of these cells will be carried out by 4C-seq, RNA-seq, enhancer assays, etc.
Group Members

Oktay Lab. on Neuro-Genomics
Research Group Leader
Yavuz OKTAY
yavuz.oktay@ibg.edu.tr
+90 232 299 41 00
(5141)
+9
0 232 299 41 64

Ayşe Semra HIZ
Research Group Member
semra.hiz@ibg.edu.tr

Pınar GENÇPINAR
PhD Student
pinar.gencpinar@ibg.edu.tr

Ayşe İpek POLAT
PhD Student
ayseipek.polat@ibg.edu.tr

Aykut KURUOĞLU
PhD Student
aykut.kuruoglu@ibg.edu.tr

Ayşe Semra HIZ
PhD Student
semra.hiz@ibg.edu.tr

Ece SÖNMEZLER
PhD Student
ece.sonmezler@ibg.edu.tr

Güneş Şayan CAN
PhD Student
gunes.can@ibg.edu.tr

Ebru BAKIR
Researcher
ebru.bakir@ibg.edu.tr

Şimal KAYIKÇI
MSc Student
simal.kayikci@ibg.edu.tr

Heval ŞEKER
Undergraduate Student
heval.seker@ibg.edu.tr

Delfin ALPSOY
MSc Student
delfin.alpsoy@ibg.edu.tr

Bilgesu ÖZTÜRK
PhD Student
bilgesu.genc@ibg.edu.tr

Dilara GÜNGÖR
MSc Student
dilara.gungor@std.ibg.edu.tr

Sevcan MERCAN
Visiting Researcher
sevcan.mercan@msfr.ibg.edu.tr
Former Members

Tutku YARAŞ
Research Technician
tutku.yaras@ibg.edu.tr

Ebru BAKIR
MSc Student
ebru.bakir@ibg.edu.tr

Kaan OKAY
MSc Student
kaan.okay@msfr.ibg.edu.tr

Ece SÖNMEZLER
MSc Student
ece.sonmezler@ibg.edu.tr

Fadime ÖZTOPRAK
PhD Student
fadime.oztoprak@ibg.edu.tr

Melike TETİK
Visiting Researcher
melike.tetik@ibg.edu.tr

Burcu EKİNCİ
PhD Student
burcu.ekinci@ibg.edu.tr

Işık Esin KIROĞLU
MSc Student
isikesin.kiroglu@ibg.edu.tr

Tutku YARAŞ
PhD Student
tutku.yaras@ibg.edu.tr

Özgem UYSAL
Undergraduate Student
ozgem.uysal@msfr.ibg.edu.tr

BEYZA NUR BİLGİLİ
Undergraduate Student
beyzanur.bilgili@msfr.ibg.edu.tr

Zeynep BİLGETEKİN
Undergraduate Student
None

Esra BAŞKURT
MSc Student
esra.baskurt@msfr.ibg.edu.tr

AHMED HASSAN MOHAMED HUSSEIN IBRAHIM AHMED HASSAN MOHAMED HUSSEIN IBRAHIM
PhD Student
ahmed.ibrahim@ibg.edu.tr

Talip ZENGİN
PhD Student
talip.zengin@ibg.edu.tr

Aleyna Dilan KIRAN
Undergraduate Student
aleyna.kiran@msfr.ibg.edu.tr

Berfin DEMİRÖZER
Researcher
berfin.demirozer@ibg.edu.tr

Bilgesu ÖZTÜRK
PhD Student
bilgesu.genc@ibg.edu.tr

Emir ARSLAN
Undergraduate Student
emir.arslan@std.ibg.edu.tr

Elif ALTUNKAYNAK
Undergraduate Student
elif.altunkaynak@std.ibg.edu.tr
News
Our lab has become part of newly established MRC-ICGNMD center on neuromuscular diseases
International Centre for Genomic Medicine in Neuromuscular Diseases (ICGNMD) has just been established and awarded 5-year funding by the Medical Research Council (MRC)-UK. The centre brings together clinicians and researchers from UK, Turkey, India, South Africa, Brazil and Zambia to advance genomic medicine in neuromuscular diseases. The inauguration ceremony of the centre took place on April 3, 2019 at House of Lords in London, with attendance by Turkish counsellor and Prof. Lord Kakkar among the audience.
Selected Publications
Can GŞ, Bakır E, Oktay Y. Functional Annotation of Bipolar Disorder 2 Risk Location Implicates Novel Susceptibility Genes.. Neuropsychobiology. 2025 January : 1-12. doi:10.1159/000543504. Download
Ak B, Akısü M, Durmaz A, Yalaz M, Terek D, Sönmezler E, Oktay Y, Akın H, Aykut A. Expanding the genetic spectrum of short rib polydactyly syndrome: Novel DYNC2H1 variants and functional insights.. Bone. 2025 August ; 197 : 117511. doi:10.1016/j.bone.2025.117511. Download
Yaraş T, Oktay Y, Karakülah G. PGSXplorer: an integrated nextflow pipeline for comprehensive quality control and polygenic score model development.. PeerJ. 2025 February ; 13 : e18973. doi:10.7717/peerj.18973. Download
Durmus S, Yucesan E, Aktug S, Utz B, Caglayan AO, Gencpinar P, Günay C, Oktay Y, Yildirim RN, Yigit A, Ozbek U. Management of rare and undiagnosed diseases: insights from researchers and healthcare professionals in Türkiye.. Frontiers in public health. 2025 January ; 12 : 1501942. doi:10.3389/fpubh.2024.1501942. Download
Cark O, Katkat E, Aydogdu I, Iscan E, Oktay Y, Ozhan G. tubg1 Somatic Mutants Show Tubulinopathy-Associated Neurodevelopmental Phenotypes in a Zebrafish Model.. Molecular neurobiology. 2024 August . doi:10.1007/s12035-024-04448-2. Download
Falabella M, Pizzamiglio C, Tabara LC, Munro B, Abdel-Hamid MS, Sonmezler E, Macken WL, Lu S, Tilokani L, Flannery PJ, Patel N, Pope SAS, Heales SJR, Hammadi DBH, Alston CL, Taylor RW, Lochmuller H, Woodward CE, Labrum R, Vandrovcova J, Houlden H, Chronopoulou E, Pierre G, Maroofian R, Hanna MG, Taanman JW, Hiz S, Oktay Y, Zaki MS, Horvath R, Prudent J, Pitceathly RDS. Biallelic PTPMT1 variants disrupt cardiolipin metabolism and lead to a neurodevelopmental syndrome.. Brain : a journal of neurology. 2024 August . doi:10.1093/brain/awae268. Download
Ülgen E, Gerlevik U, Gerlevik S, Oktay Y, Sezerman OU, Turcan Ş, Ozduman K. A microdeletion event at 19q13.43 in IDH-mutant astrocytomas is strongly correlated with MYC overexpression.. Acta neuropathologica communications. 2024 June ; 12 (1) : 95. doi:10.1186/s40478-024-01811-1. Download
Köse SN, Yaraş T, Bursali A, Oktay Y, Yandim C, Karakülah G. Expressions of the satellite repeat HSAT5 and transposable elements are implicated in disease progression and survival in glioma.. Turkish journal of biology = Turk biyoloji dergisi. 2024 July ; 48 (4) : 242-256. doi:10.55730/1300-0152.2700. Download
Cavusoglu D, Ozturk G, Turkdogan D, Kurul SH, Yis U, Komur M, Incecik F, Kara B, Sahin T, Unver O, Dilber C, Mert GG, Gunay C, Uzan GS, Ersoy O, Oktay Y, Mermer S, Tuncer GO, Gungor O, Ozcora GDK, Gumus U, Sezer O, Cetin GO, Demir F, Yilmaz A, Gurbuz G, Topcu M, Topaloglu H, Ceylan AC, Ceylaner S, Gleeson JG, Icagasioglu DF, Sonmez FM. Evaluation of the Patients with the Diagnosis of Pontocerebellar Hypoplasia: A Multicenter National Study.. Cerebellum (London, England). 2024 October ; 23 (5) : 1950-1965. doi:10.1007/s12311-024-01690-1. Download
Sönmezler E, Stuani C, Hız Kurul S, Güngör S, Buratti E, Oktay Y. Characterization and Engineered U1 snRNA Rescue of Splicing Variants in a Turkish Neurodevelopmental Disease Cohort.. Human mutation. 2024 May ; 2024 : 7760556. doi:10.1155/2024/7760556. Download
Özbek M, Toy HI, Oktay Y, Karakülah G, Suner A, Pavlopoulou A. An in silico approach to the identification of diagnostic and prognostic markers in low-grade gliomas.. PeerJ. 2023 March ; 11 : e15096. doi:10.7717/peerj.15096. Download
Tetik M, Direk N, Uzgan BÖ, Aykaç C, Ekinci B, Yaraş T, Kuruoğlu A, Özel F, Ermiş Ç, Alkın T, Oktay Y. Associations Between Blood Levels of NLRP3 Inflammasome Components and Obsessive Compulsive Disorder.. Noro psikiyatri arsivi. 2023 February ; 60 (1) : 28-36. doi:10.29399/npa.28127. Download
Lecca M, Pehlivan D, Suñer DH, Weiss K, Coste T, Zweier M, Oktay Y, Danial-Farran N, Rosti V, Bonasoni MP, Malara A, Contrò G, Zuntini R, Pollazzon M, Pascarella R, Neri A, Fusco C, Marafi D, Mitani T, Posey JE, Bayramoglu SE, Gezdirici A, Hernandez-Rodriguez J, Cladera EA, Miravet E, Roldan-Busto J, Ruiz MA, Bauzá CV, Ben-Sira L, Sigaudy S, Begemann A, Unger S, Güngör S, Hiz S, Sonmezler E, Zehavi Y, Jerdev M, Balduini A, Zuffardi O, Horvath R, Lochmüller H, Rauch A, Garavelli L, Tournier-Lasserve E, Spiegel R, Lupski JR, Errichiello E. Bi-allelic variants in the ESAM tight-junction gene cause a neurodevelopmental disorder associated with fetal intracranial hemorrhage.. American journal of human genetics. 2023 April ; 110 (4) : 681-690. doi:10.1016/j.ajhg.2023.03.005. Download
Ozturk BT, Ozel F, Yaras T, Ekinci B, Oktay Y, Aysevener EO, Alkın T, Direk N. The Relationship Between Cardiovascular Disease Risk and Major Depression.. Noro psikiyatri arsivi. 2023 May ; 60 (2) : 124-128. doi:10.29399/npa.28191. Download
Günay Ç, Aykol D, Özsoy Ö, Sönmezler E, Hanci YS, Kara B, Akkoyunlu Sünnetçi D, Cine N, Deniz A, Özer T, Ölçülü CB, Yilmaz Ö, Kanmaz S, Yilmaz S, Tekgül H, Yildiz N, Acar Arslan E, Cansu A, Olgaç Dündar N, Kusgoz F, Didinmez E, Gençpinar P, Aksu Uzunhan T, Ertürk B, Gezdirici A, Ayaz A, Ölmez A, Ayanoğlu M, Tosun A, Topçu Y, Kiliç B, Aydin K, Çağlar E, Ersoy Kosvali Ö, Okuyaz Ç, Besen Ş, Tekin Orgun L, Erol İ, Yüksel D, Sezer A, Atasoy E, Toprak Ü, Güngör S, Ozgor B, Karadağ M, Dilber C, Şahinoğlu B, Uyur Yalçin E, Eldes Hacifazlioglu N, Yaramiş A, Edem P, Gezici Tekin H, Yilmaz Ü, Ünalp A, Turay S, Biçer D, Gül Mert G, Dokurel Çetin İ, Kirik S, Öztürk G, Karal Y, Sanri A, Aksoy A, Polat M, Özgün N, Soydemir D, Sarikaya Uzan G, Ülker Üstebay D, Gök A, Yeşilmen MC, Yiş U, Karakülah G, Bursali A, Oktay Y, Hiz Kurul S. Shared Biological Pathways and Processes in Patients with Intellectual Disability: A Multicenter Study.. Neuropediatrics. 2023 August ; 54 (4) : 225-238. doi:10.1055/a-2034-8528. Download
Özsoy Ö, Cinleti T, Günay Ç, Sarıkaya Uzan G, Yeşilmen MC, Lochmüller H, Horvath R, Yiş U, Oktay Y, Hiz Kurul S. DPAGT1-CDG: Report of Two New Pediatric Patients and Brief Review of the Literature.. Molecular syndromology. 2023 August ; 14 (4) : 322-330. doi:10.1159/000529494. Download
Wilson LA, Macken WL, Perry LD, Record CJ, Schon KR, Frezatti RSS, Raga S, Naidu K, Köken ÖY, Polat I, Kapapa MM, Dominik N, Efthymiou S, Morsy H, Nel M, Fassad MR, Gao F, Patel K, Schoonen M, Bisschoff M, Vorster A, Jonvik H, Human R, Lubbe E, Nonyane M, Vengalil S, Nashi S, Srivastava K, Lemmers RJLF, Reyaz A, Mishra R, Töpf A, Trainor CI, Steyn EC, Mahungu AC, van der Vliet PJ, Ceylan AC, Hiz AS, Çavdarlı B, Semerci Gündüz CN, Ceylan GG, Nagappa M, Tallapaka KB, Govindaraj P, van der Maarel SM, Narayanappa G, Nandeesh BN, Wa Somwe S, Bearden DR, Kvalsund MP, Ramdharry GM, Oktay Y, Yiş U, Topaloğlu H, Sarkozy A, Bugiardini E, Henning F, Wilmshurst JM, Heckmann JM, McFarland R, Taylor RW, Smuts I, van der Westhuizen FH, Sobreira CFDR, Tomaselli PJ, Marques W Jr, Bhatia R, Dalal A, Srivastava MVP, Yareeda S, Nalini A, Vishnu VY, Thangaraj K, Straub V, Horvath R, Chinnery PF, Pitceathly RDS, Muntoni F, Houlden H, Vandrovcova J, Reilly MM, Hanna MG. Neuromuscular disease genetics in under-represented populations: increasing data diversity.. Brain : a journal of neurology. 2023 December ; 146 (12) : 5098-5109. doi:10.1093/brain/awad254. Download
Leman Binokay, Yavuz Oktay & Gökhan Karakülah. An API for dynamic estimation of reference intervals for functional abundances of gut microbiota. Biologia. 2023 October ; 79 : 343-353. doi:10.1007/s11756-023-01556-7. Download
Koçhan N, Oktay Y, Karakülah G. StemnesScoRe: an R package to estimate the stemness of glioma cancer cells at single-cell resolution.. Turkish journal of biology = Turk biyoloji dergisi. 2023 December ; 47 (6) : 383-392. doi:10.55730/1300-0152.2672. Download
Harwood AJ, Petrakis S, Oktay Y, Pasterkamp RJ. Editorial: Mental health: cell models to mechanisms.. Frontiers in cell and developmental biology. 2023 July ; 11 : 1244425. doi:10.3389/fcell.2023.1244425. Download
Martin Kuiper, Joseph Bonello, Jesualdo T.Fernández-Breis, PhilippBucher, Matthias E.Futschik, Pascale Gaudet, Ivan V. Kulakovskiy, Luana Licata, Colin Logie, Ruth C.Lovering, Vsevolod J.Makeev, Sandra Orchard, Simona Panni, Livia Perfetto, David Santo, Stefan Schulz, Steven Vercruyssea, Daniel R. Zerbino, Astrid Lægreid, The GRECO Consortium. The gene regulation knowledge commons: the action area of GREEKC. Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms. 2022 January ; 1865 (1) : 194768. doi:10.1016/j.bbagrm.2021.194768. Download
Okay K, Varış PÜ, Miral S, Pavlopoulou A, Oktay Y, Karakülah G. Whole Genome Analysis of Dizygotic Twins With Autism Reveals Prevalent Transposon Insertion Within Neuronal Regulatory Elements: Potential Implications for Disease Etiology and Clinical Assessment.. Journal of autism and developmental disorders. 2022 June . doi:10.1007/s10803-022-05636-6. Download
Engin Köse 1, Elçin Çağatay 2, Tutku Yaraş 3, Pelin Teke Kısa 4, Seminay Güler 5, Zümrüt Arslan Gülten 4, Mesut Akarsu 5, Yavuz Oktay 6, Hülya Ayar Kayalı 7, Nur Arslan 4. Could lysosomal acid lipase enzyme activity be used for clinical follow-up in cryptogenic cirrhosis?. Turkish Journal of Medical Sciences. 2022 August ; 52 (4) : 1075-1084. doi:10.55730/1300-0144.5410. Download
Arlt A, Kohlschmidt N, Hentschel A, Bartels E, Groß C, Töpf A, Edem P, Szabo N, Sickmann A, Meyer N, Schara-Schmidt U, Lau J, Lochmüller H, Horvath R, Oktay Y, Roos A, Hiz S. Novel insights into PORCN mutations, associated phenotypes and pathophysiological aspects.. Orphanet journal of rare diseases. 2022 January ; 17 (1) : 29. doi:10.1186/s13023-021-02068-w. Download
Pizzamiglio C, Pitceathly RDS, Lunn MP, Brady S, De Marchi F, Galan L, Heckmann JM, Horga A, Molnar MJ, Oliveira ASB, Pinto WBVR, Primiano G, Santos E, Schoser B, Servidei S, Sgobbi Souza PV, Venugopalan V, Hanna MG, Dimachkie MM, Machado PM, Neuromuscular Diseases and COVID-19 Study Group. Factors associated with the severity of COVID-19 outcomes in people with neuromuscular diseases: Data from the International Neuromuscular COVID-19 Registry.. European journal of neurology. 2022 October . doi:10.1111/ene.15613. Download
Doğa Eskier, Evren Akalp, Özlem Dalan, Gökhan Karakülah, Yavuz Oktay. Current mutatome of SARS-CoV-2 in Turkey reveals mutations of interest. Turkish Journal of Biology. 2021 February ; 45 (1) : 104-113. doi:10.3906/biy-2008-56. Download
Koçhan N, Eskier D, Suner A, Karakülah G, Oktay Y. Different selection dynamics of S and RdRp between SARS-CoV-2 genomes with and without the dominant mutations.. Infection, genetics and evolution : journal of molecular epidemiology and evolutionary genetics in infectious diseases. 2021 July ; 91 : 104796. doi:10.1016/j.meegid.2021.104796. Download
Okay K, Varış PÜ, Miral S, Ekinci B, Yaraş T, Karakülah G, Oktay Y. Alternative splicing and gene co-expression network-based analysis of dizygotic twins with autism-spectrum disorder and their parents.. Genomics. 2021 July ; 113 (4) : 2561-2571. doi:10.1016/j.ygeno.2021.05.038. Download
Serdal Gungor, Yavuz Oktay, Semra Hiz, Álvaro Aranguren-Ibáñez, Ipek Kalafatcilar, Ahmet Yaramis, Ezgi Karaca, Uluc Yis, Ece Sonmezler, Burcu Ekinci, Mahmut Aslan, Elmasnur Yilmaz, Sunitha Balaraju, Nora Szabo, Steven Laurie, Sergi Beltran, Daniel G. MacArthur, Denisa Hathazi, Rita Horvath. Autosomal recessive variants in TUBGCP2 alter the γ-tubulin ring complex leading to neurodevelopmental disease. iScience. 2021 January ; 24 (1) . doi:10.1016/j.isci.2020.101948. Download
Kurul SH, Oktay Y, Töpf A, Szabó NZ, Güngör S, Yaramis A, Sonmezler E, Matalonga L, Yis U, Schon K, Paramonov I, Kalafatcilar İP, Gao F, Rieger A, Arslan N, Yilmaz E, Ekinci B, Edem PP, Aslan M, Özgör B, Lochmüller A, Nair A, O'Heir E, Lovgren AK, Broad Center for Mendelian Genomics., Maroofian R, Houlden H, Polavarapu K, Roos A, Müller JS, Hathazi D, Chinnery PF, Laurie S, Beltran S, Lochmüller H, Horvath R. High diagnostic rate of trio exome sequencing in consanguineous families with neurogenetic diseases.. Brain : a journal of neurology. 2021 November . doi:10.1093/brain/awab395. Download
Richard EM, Bakhtiari S, Marsh APL, Kaiyrzhanov R, Wagner M, Shetty S, Pagnozzi A, Nordlie SM, Guida BS, Cornejo P, Magee H, Liu J, Norton BY, Webster RI, Worgan L, Hakonarson H, Li J, Guo Y, Jain M, Blesson A, Rodan LH, Abbott MA, Comi A, Cohen JS, Alhaddad B, Meitinger T, Lenz D, Ziegler A, Kotzaeridou U, Brunet T, Chassevent A, Smith-Hicks C, Ekstein J, Weiden T, Hahn A, Zharkinbekova N, Turnpenny P, Tucci A, Yelton M, Horvath R, Gungor S, Hiz S, Oktay Y, Lochmuller H, Zollino M, Morleo M, Marangi G, Nigro V, Torella A, Pinelli M, Amenta S, Husain RA, Grossmann B, Rapp M, Steen C, Marquardt I, Grimmel M, Grasshoff U, Korenke GC, Owczarek-Lipska M, Neidhardt J, Radio FC, Mancini C, Claps Sepulveda DJ, McWalter K, Begtrup A, Crunk A, Guillen Sacoto MJ, Person R, Schnur RE, Mancardi MM, Kreuder F, Striano P, Zara F, Chung WK, Marks WA, van Eyk CL, Webber DL, Corbett MA, Harper K, Berry JG, MacLennan AH, Gecz J, Tartaglia M, Salpietro V, Christodoulou J, Kaslin J, Padilla-Lopez S, Bilguvar K, Munchau A, Ahmed ZM, Hufnagel RB, Fahey MC, Maroofian R, Houlden H, Sticht H, Mane SM, Rad A, Vona B, Jin SC, Haack TB, Makowski C, Hirsch Y, Riazuddin S, Kruer MC. Bi-allelic variants in SPATA5L1 lead to intellectual disability, spastic-dystonic cerebral palsy, epilepsy, and hearing loss.. American journal of human genetics. 2021 October ; 108 (10) : 2006-2016. doi:10.1016/j.ajhg.2021.08.003. Download
Ana Töpf, Yavuz Oktay, Sunitha Balaraju, Elmasnur Yilmaz, Ece Sonmezler, Uluc Yis, Steven Laurie, Rachel Thompson, Andreas Roos, Daniel G MacArthur, .... Severe neurodevelopmental disease caused by a homozygous TLK2 variant. European journal of human genetics. 2020 March ; 28 (3) : 383-387. doi:10.1038/s41431-019-0519-x. Download
Ahmet Yaramis, Hanns Lochmüller, Ana Töpf, Ece Sonmezler, Elmasnur Yilmaz, Semra Hiz, Uluc Yis, Serdal Gungor, Ayse Ipek Polat, Pinar Edem, Sergi Beltran, Steven Laurie, Aysenur Yaramis, Rita Horvath, Yavuz Oktay. COL4A1-related autosomal recessive encephalopathy in 2 Turkish children. Neurology Genetics. 2020 January ; 6 (1) : 1. doi:10.1212/NXG.0000000000000392. Download
Melike Tetik, Nese Direk, Betul Onder, Cansu Aykac, Burcu Ekinci, Tutku Yaras, Aykut Kuruoglu, Cagatay Ermis, Tunc Alkin, Yavuz Oktay. Blood Levels of NLRP3 Inflammasome Components is Associated with Obsessive Compulsive Disorder. Biological Psychiatry. 2020 January . doi:10.1101/2020.01.30.20019646. Download
Doğa Eskier, Gökhan Karakülah, Aslı Suner, Yavuz Oktay. RdRp mutations are associated with SARS-CoV-2 genome evolution. BioRxiv. 2020 May . doi:10.1101/2020.05.20.104885. Download
Eskier D, Karakülah G, Suner A, Oktay Y. RdRp mutations are associated with SARS-CoV-2 genome evolution.. PeerJ. 2020 July ; 8 : e9587. doi:10.7717/peerj.9587. Download
Oktay Y, Güngör S, Zeltner L, Wiethoff S, Schöls L, Sonmezler E, Yilmaz E, Munro B, Bender B, Kernstock C, Kaemereit S, Liepelt I, Töpf A, Yis U, Laurie S, Yaramis A, Zuchner S, Hiz S, Lochmüller H, Schüle R, Horvath R. Confirmation of TACO1 as a Leigh Syndrome Disease Gene in Two Additional Families.. Journal of neuromuscular diseases. 2020 June ; 7 (3) : 301-308. doi:10.3233/JND-200510. Download
Paketci C , Edem P , Hiz S , Sonmezler E , Soydemir D , Sarikaya Uzan G ,Oktay Y, O'Heir E , Beltran S, Laurie S, Töpf A, Lochmuller H, Horvath R, Yis U. Successful treatment of intractable epilepsy with ketogenic diet therapy in twins with ALG3-CDG.. Brain and Development. 2020 August ; 42 (7) : 539-545. doi:10.1016/j.braindev.2020.04.008. Download
Eskier D, Suner A, Karakülah G, Oktay Y. Mutation density changes in SARS-CoV-2 are related to the pandemic stage but to a lesser extent in the dominant strain with mutations in spike and RdRp. PeerJ. 2020 June ; 8 : e9703. doi:10.7717/peerj.9703. Download
Eskier D, Suner A, Oktay Y, Karakülah G. Mutations of SARS-CoV-2 nsp14 exhibit strong association with increased genome-wide mutation load.. PeerJ. 2020 October ; 8 : e10181. doi:10.7717/peerj.10181. Download
Ülgen E, Karacan S, Gerlevik U, Can Ö, Bilguvar K, Oktay Y, B Akyerli C, K Yüksel Ş, E Danyeli A, Tihan T, Sezerman OU, Yakıcıer MC, Pamir MN, Özduman K. Mutations and Copy Number Alterations in IDH Wild-Type Glioblastomas Are Shaped by Different Oncogenic Mechanisms. Biomedicines. 2020 December ; 8 (12) . doi:10.3390/biomedicines8120574. Download
Yiş U., Hız S., Güneş S., Diniz G., Sönmezler E., Yılmaz E., Oktay Y. . Dihydropyridine receptor congenital myopathy in a consanguineous Turkish family. Journal of Neuromuscular Diseases. 2019 June ; 6 (3) : 377-384. doi:10.3233/JND-190383. Download
Ege Ulgen, Ozge Can, Kaya Bilguvar, Yavuz Oktay, Cemaliye Akyerli, Ayça Danyeli, Cengiz Yakicier, Ugur Sezerman, Necmettin Pamir Koray Özduman. Whole Exome Sequencing-Based Analysis Identifies DNA Damage Repair Deficiency as a Major Contributor to Gliomagenesis in Adult Diffuse Gliomas. J Neurosurg. 2019 April : 1-12. doi:10.3171/2019.1.JNS182938. Download
Akyerli CB, Yüksel Ş, Can Ş, Erson-Omay EZ, Oktay Y, Coşgun E, Ülgen E, Erdemgil Y, Sav A, von Deimling A, Günel M, Yakıcıer MC, Pamir MN, Öz............ Use of telomerase promoter mutations to mark specific molecular subsets with reciprocal clinical behavior in IDH mutant and IDH wild-type diffuse gliomas. J Neurosurg. 2018 April ; 128 (4) : 1102-1114. doi:10.3171/2016.11.JNS16973. Download
Oktay Y., Boylu C.A., Özduman K.. Gliom gelişiminde genetik yatkınlığın rolü. Türk Nöroşir Derg. 2017 March ; 27 (2) : 122-130. doi: . Download
Pavlopoulou, Athanasia; Oktay, Yavuz; Vougas, Konstantinos; Louka, Maria; Vorgias, Constantinos E.; Georgakilas, Alexandros G.. Determinants of resistance to chemotherapy and ionizing radiation in breast cancer stem cells. CANCER LETTERS. 2016 October ; 380 (2) : 485-493. doi:10.1016/j.canlet.2016.07.018. Download
Oktay, Yavuz; Ulgen, Ege; Can, Ozge; Akyerli, Cemaliye B.; Yuksel, Sirin; Erdemgil, Yigit; Durasi, I. Melis; Henegariu, Octavian Ioan; Nanni, E. Paolo; Selevsek, Nathalie; Grossmann, Jonas; Erson-Omay, E. Zeynep; Bai, Hanwen; Gupta, Manu; Lee, William; Turcan, Sevin; Ozpinar, Aysel; Huse, Jason T.; Sav, M. Aydin; Flanagan, Adrienne; Gunel, Murat; Sezerman, O. Ugur; Yakicier, M. Cengiz; Pamir, M. Necmettin; Ozduman, Koray. IDH-mutant glioma specific association of rs55705857 located at 8q24.21 involves MYC deregulation. SCIENTIFIC REPORTS. 2016 June ; 6 . doi:10.1038/srep27569. Download
Total : 47
Selected Book Chapters
Genomik ve Uygulamaları (2023). Moleküler Biyoloji ve Genetik (Sağlık Alanında ve Biyoteknolojide Uygulamalar). TÜBA.
Genes and Genomes, The Nucleus (2016). The Cell: A Molecular Approach Seventh Edition. Oxford University Press.
Chapter 3: Principles of Medical Genetics, Chromosomal Disorders and Single-Gene Inheritance (2015). Current Applications of Biotechnology. Erciyes Universitesi.
Total : 3
Projects
Medical Research Council (MRC), UK - RD : MRC Strategic Award for an International Centre for Genomic Medicine in Neuromuscular Diseases, Finished
The Scientific and Technological Research Council of Turkey - TUBITAK - RD : Using Colocalization Approach for Epigenomic Analysis of Lithium Response-Related Genomic Loci in Bipolar Disorder, Ongoing
The Scientific and Technological Research Council of Turkey - TUBITAK - RD : Evaluation of the Corrective Effects of Small Molecules Identified by Multi-Omic Analyses on Mitochondrial Defects Due to PTPMT1 Deficiency in PTPMT1 Knockout Neural Progenitor Cells and Zebrafish Models, Funded, Not Started
European Union - RD : The European Rare Disease Research Alliance (ERDERA), Ongoing
European Union - RD : Genome of Europe (GoE), Ongoing
Awards
- MRC Strategic Award for an International Centre for Genomic Medicine in Neuromuscular Diseases by Medical Research Council (MRC, UK), 2018
- Young Investigator (GEBİP) Awards by Turkish Academy of Sciences (TÜBA), 2017
- Young Scientist (BAGEP) Award by Science Academy, 2013
Academic Memberships
- The Science Academy, Turkey, 2013
- TÜBA-Turkish Academy Of Science, 2017
Contact

Oktay Lab. on Neuro-Genomics
Research Group Leader
Yavuz OKTAY
yavuz.oktay@ibg.edu.tr
+90 232 299 41 00
(5141)
+9
0 232 299 41 64
