Emerging Viral Diseases
Principal Investigator: Assist. Prof. Zeynep A. KOÇER
Since the 1918 Spanish Flu pandemic, influenza A viruses have caused four pandemics, resulting in millions of deaths. Although wild waterfowl serve as their primary natural reservoir, these viruses are capable of infecting a wide range of hosts. Owing to genetic changes they undergo, influenza viruses can acquire the ability to transmit both intra- and inter-species, thereby enabling them to cause epidemics and pandemics. Therefore, monitoring changes in the viral genome and investigating the potential effects of these changes through computational and experimental approaches not only allows for a better understanding of viruses that pose a pandemic risk but also contributes to reducing the impact of potential future outbreaks.
In our laboratory, we perform genome analyses and molecular characterizations of viral pathogens that cause infections in humans, particularly influenza viruses, and investigate viral evolution, ecology, and risk assessment through bioinformatics/computational analyses. Additionally, preliminary research related to the development of antivirals and vaccines for the treatment and prevention of viral diseases is among our laboratory’s primary research areas.
Furthermore, software for bioinformatics-based risk assessment tools is being developed by our laboratory to enable the prediction of how viral and host factors contribute to the transition of avian influenza viruses from low to high pathogenicity.
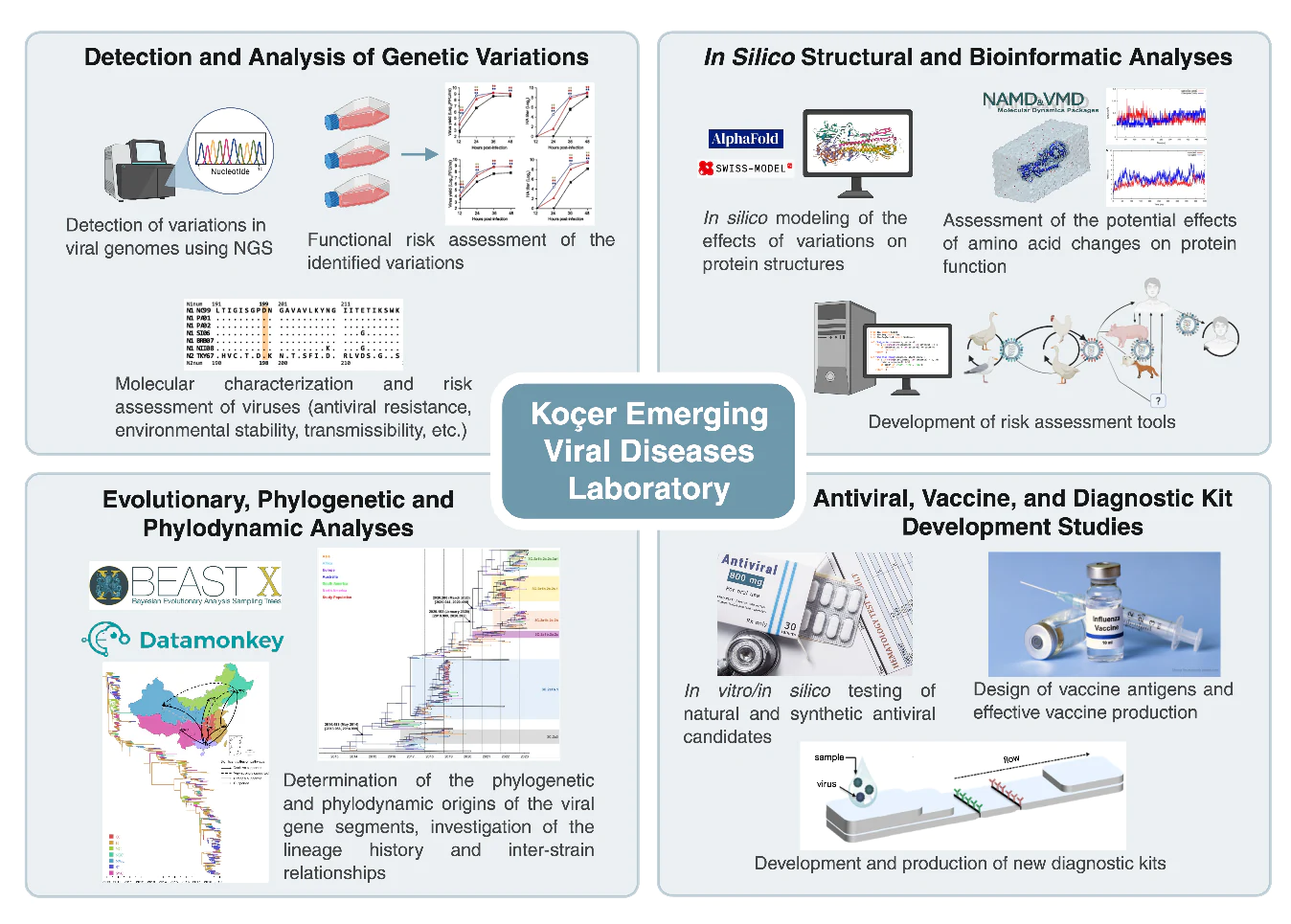
Research Areas
Bioinformatic Analysis
NGS viral genome sequenceanalysis
Phylogenetic and phylodynamic analysis
Molecular dynamics simulations
Development of risk assessment tools
Transcriptomic analysis of host-pathogen interactions
Identification of Antiviral Candidate Molecules
In vitro efficacy tests of antiviral candidate plant extracts
In silico and in vitro efficacy testing of antiviral candidate small molecules
Vaccine Development
Vaccine development using reverse genetics methods
Publications
Azbazdar, M. E., Dikmenogullari, M., Kavalci, Z., & Koçer, Z. A. (2025). Genetic Evolution of the Hemagglutinin Genes of Seasonal Influenza A Viruses in Türkiye Between 2017 and 2023. Influenza and other respiratory viruses, 19(7), e70134. https://doi.org/10.1111/irv.70134
M. E. Azbazdarand Z. A. Koçer (2025). "İnfluenza B VirüslerininEpidemiyolojisindeDemografi, AntijenikÖzelliklerve Filogenetik İlişkilerinBağlantısı," Türk MikrobiyolojiCemiyetiDergisi, vol.55, no.1, pp.73-92, 2025. doi:10.54453/tmcd.2025.42744
Bai, Y., Lei, H., Song, W., Shin, S. C., Wang, J., Xiao, B., Koçer, Z. A., Song, M. S., Webster, R., Webby, R. J., Wong, S. S., & Zanin, M. (2024). Amino acids in the polymerase complex of shorebird-isolated H1N1 influenza virus impact replication and host-virus interactions in mammalian models. Emerging microbes & infections, 13(1), 2332652. https://doi.org/10.1080/22221751.2024.2332652
Azbazdar, M. E., Akıncı, G., Güler, E., Koçer, Z. A. (2022). The effect of sediment composition and polyethylene glycol precipitation on the detection limit of H6N2 influenza virus in sediment samples. Marine and Life Sciences, 4(1), 53-62. https://doi.org/10.51756/marlife.1091169
Mercan, Y., Atim, G., Kayed, A. E., Azbazdar, M. E., Kandeil, A., Ali, M. A., Rubrum, A., McKenzie, P., Webby, R. J., Erima, B., Wabwire-Mangen, F., Ukuli, Q. A., Tugume, T., Byarugaba, D. K., Kayali, G., Ducatez, M. F., & Koçer, Z. A. (2021). Molecular Characterization of Closely Related H6N2 Avian Influenza Viruses Isolated from Turkey, Egypt, and Uganda. Viruses, 13(4), 607. https://doi.org/10.3390/v13040607
Projects
Ongoing Projects
- TÜBİTAK-1001: Investigation of Antiviral Effects of Small Molecules Targeting NS2/NEP Protein of Influenza Viruses and Interaction Interfaces with Viral/Host Proteins via in silico and in vitro Approaches, Funded, Not Started (Coordinator)
- EUPAHW Call: Evolution of avian influenza viruses in the context of vaccination (EVIVA), Funded, Not Started (Coordinator)
- TÜBİTAK - NSF Call:Genetic and Phenotypic Contribution of Swine Influenza Viruses to the Ecological and Evolutionary Dynamics of Human Influenza A Viruses (FLU-SHIFT), Ongoing(Coordinator)
- ERA-NETCofund ICRAD Call: Identification of factors driving the emergence and spread of avian influenza viruses with zoonotic potential (FLU-SWITCH), Ongoing(Coordinator)
- TÜBİTAK - 1004: Prevention and Therapeutics National Platform, Development of biotechnological vaccine formulations for H9N2 avian influenza, Ongoing (Coordinator, Project 7)
- TÜBİTAK - 1002: The investigation of the antiviral effect of Cistus laurifolius plantextract, rich in phenolic compounds, against seasonal influenza A viruses, Ongoing (Coordinator)
- HacettepeUniversity -BAP: Investigation of Perfluorooctane Sulfonic Acid and Perfluorooctanoic Acid Exposure on the Cell Damage and Entry of pseudo-SARS-CoV-2 into Cells in A549-hACE2-TMPRSS2 Cell Line; Interleukin-17 Signaling and Severity of Lung Damage Caused by SARS-CoV-2 in Balb/c Mice, Ongoing (Researcher)
- BoğaziçiUniversity - BAP: The Investigation of Bat Related Blood Bacteria Prevalence in Türkiye using Genetic Methods, Ongoing (Researcher)
Completed Projects
- Dokuz Eylul University-BAP: Preliminary Study for the Optimization and Detection of Zoonotic Virus Profile thatare Carried by Bats in Türkiye using Next Generation Sequencing, Finished (Coordinator)
- Dokuz Eylul University-BAP: Assessing the Contribution of Genetic Variations and Reassortment Events thatOccurred in Influenza Viruses During 2018-2019 and 2019-2020 Seasons in AegeanRegion, Turkey, Finished (Coordinator)
- TÜBİTAK - 1001: Development of Inactive Trivalent Flu Vaccine Formulation, Finished (Researcher)
- TÜBİTAK - 1001: Assessment of Pandemic Risk for Influenza Viruses in Shorebirds via Molecular Approach, Finished (Coordinator)
- Dokuz Eylul University-BAP:Prevalence and Diversity ofInfluenza A Viruses in Sediment Samples in Gediz Delta, Finished(Coordinator)
- TÜBİTAK – 2232: Prevalence and Diversity of Influenza A Viruses in Wetland Sediments, Finished (Coordinator)
Staff
Active Members
Dr. Mukesh Nitin (Post-Doctoral Researcher)
M. Ekin Azbazdar, MSc (PhD Candidate)
Mert Dikmenoğulları, MSc (PhD Candidate)
Zeynep Kavalcı, MSc (PhD Candidate)
Nidanur Işın (MSc Student)
İlhan Mert Özalp (MSc Student)
Zeynep Uzuntuna (MSc Student)
Former Members:
Yavuz Mercan, MSc
Öykü Durak, MSc
Emre Mert Asar, MSc