Computational Structural Biology
Principal Investigator: Assist. Prof. Ezgi KARACA
Karaca Lab. focuses on understanding the structure-function relationships of biomolecular complexes using computational approaches. While experimental methods have revealed many biomolecular structures, they cannot keep pace with the rapid growth of data in fields like genomics, proteomics, and biochemistry. To bridge this gap, we leverage and develop state-of-the-art computational tools in Structural Biology. Since the breakthrough of artificial intelligence (AI)-based approaches, we’ve integrated them into our workflows to interpret omics data at the structural level more effectively.
Our research aims to provide high-accuracy structural models that offer insights into biomolecular interactions and mechanisms. By combining advanced modelling techniques with biological data, we contribute to a deeper understanding of molecular function in health and disease.
Research Areas
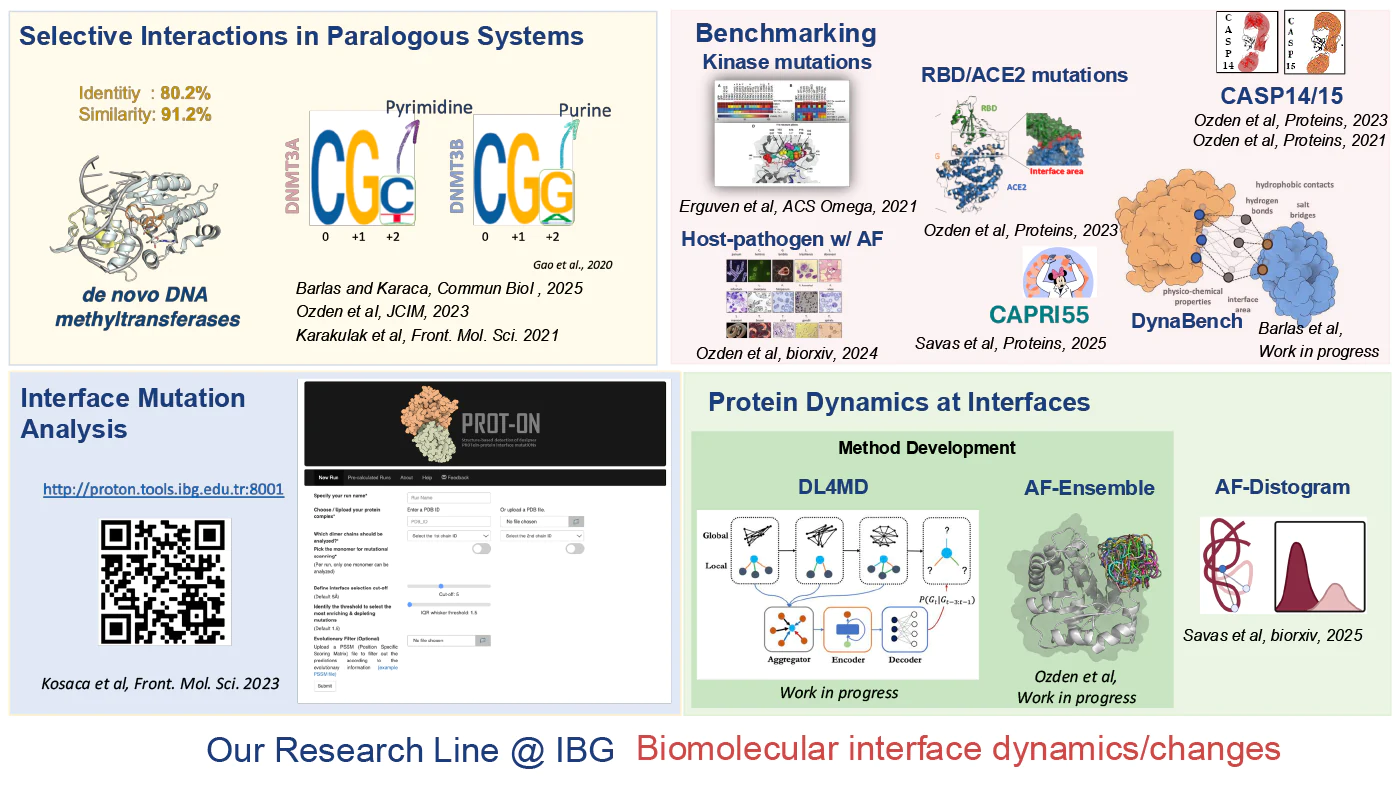
Selective Interactions in Paralog Systems: Advanced structural modeling approaches and molecular dynamics simulations are employed to explore specific interaction mechanisms between highly similar proteins, such as de novo DNA methyltransferases. Hence, we analyze the specificity-determining factors at protein–protein interfaces.
Effects of Interface Mutations: The structural impacts of mutations at protein–protein interfaces are investigated. Our web server, PROT-ON (http://proton.tools.ibg.edu.tr:8001/result/dae4ee4df0ec47e9a119fd512fdbfd98), allows interactive exploration of the effects of interface mutations.
Interface Characterization and Comparison Using AI- and Physics-Based Modeling Approaches: Interface characterization is performed using AI- and physics-based modeling approaches in systems such as kinase mutations, RBD-ACE2 interactions, and host–pathogen complexes. The performance and accuracy of these methods are evaluated through comparative benchmarking.
Development of Interface-Based Scoring Tools: A Python-based analysis tool, DynaBench, is being developed to provide detailed assessments of the structural and physicochemical properties of protein–protein interfaces.
Method Development for Understanding Molecular Dynamics: We are actively developing new approaches—such as DL4MD and AF-Ensemble—to model the dynamic behavior of protein-protein interfaces.
Deep Learning-Based Complex Protein Structure Prediction: A new generation, green and efficient; low-resource AI approach named MinnieFold (https://github.com/CSB-KaracaLab/MinnieFold) is being developed to model protein–protein complexes. It is tested in the CAPRI55 competition.
Evaluation of Protein Complex Modeling Approaches: We served as assessors in the CASP Assembly category during CASP14 and CASP15 to evaluate the accuracy and applicability of current modeling methods in structural biology. In this context, we analyzed protein complex prediction approaches using independent, systematic, and objective metrics.
Publications
Schildhauer, F., Ryl, P. S., Lauer, S. M., Lenz, S., Barlas, A. B., Ouzounidis, V. R., Jeffrey, K., Marcu, D., O’Reilly, F. J., Graziadei, A., Stuiver, M., Schmidt, K., Ewers, H., Spahn, C. M. T., Karaca, E., ... & Rappsilber, J. An NADH-controlled gatekeeper of ATP synthase. Molecular Cell. 2025.
Savaş, B., Yılmazbilek, İ., Özsan, A., & Karaca, E. Towards a Greener AlphaFold2 Protocol for Antibody–Antigen Modeling: Insights From CAPRI Round 55. Proteins: Structure, Function, and Bioinformatics. 2025.
Honorato, R. V., Trellet, M. E., Jiménez-García, B., Schaarschmidt, J. J., Giulini, M., Reys, V., Koukos, P. I., Rodrigues, J. P. G. L. M., Karaca, E., ... & Bonvin, A. M. The HADDOCK2. 4 web server for integrative modeling of biomolecular complexes. Nature protocols, 19(11), 3219-3241. 2024.
Ozden, B., Şamiloğlu, E., Özsan, A., Erguven, M., Yükrük, C., Koşaca, M., ... & Karaca, E.. Benchmarking the accuracy of structure‐based binding affinity predictors on Spike–ACE2 deep mutational interaction set. Proteins: Structure, Function, and Bioinformatics, 92(4), 529-539. 2024.
Ozden, B., Kryshtafovych, A., & Karaca, E.. The impact of AI‐based modeling on the accuracy of protein assembly prediction: insights from CASP15. Proteins: Structure, Function, and Bioinformatics, 91(12), 1636-1657. 2023.
Projects
EMBO Installation Grant #4421: Identification of Pioneer Transcription Factor Binding Modes to Chromatin
TÜBİTAK 2509 Türkiye - Fransa Bosphorus ikili işbirliği çağrısı: Exploring AlphaFold for Generation of Conformational Ensembles of Protein Complexes (AF-Ensemble)
Staff
Active Members
Ayşe Berçin Barlas, (Post Doc.)
Burcu Özden, (Post Doc.)
Büşra Savaş, (PhD.)
İbrahim Atakan Kubilay, (PhD.)
Buse Şahin, (MsC.)
Murat Taylan Şahin, (MsC.)
Gülce Berfin Ercan, (Undergrad.)
Former Members
Ayşe Berçin Barlas, (PhD.)
Burcu Özden, (PhD.)
Mehdi Koşaca, (MsC.)
Eda Şamiloğlu, (MsC.)
Deniz Doğan, (MsC.)
Tülay Karakulak, (MsC.)