Computational Sequence Analysis
Principal Investigator: Prof. Dr. Gökhan KARAKÜLAH
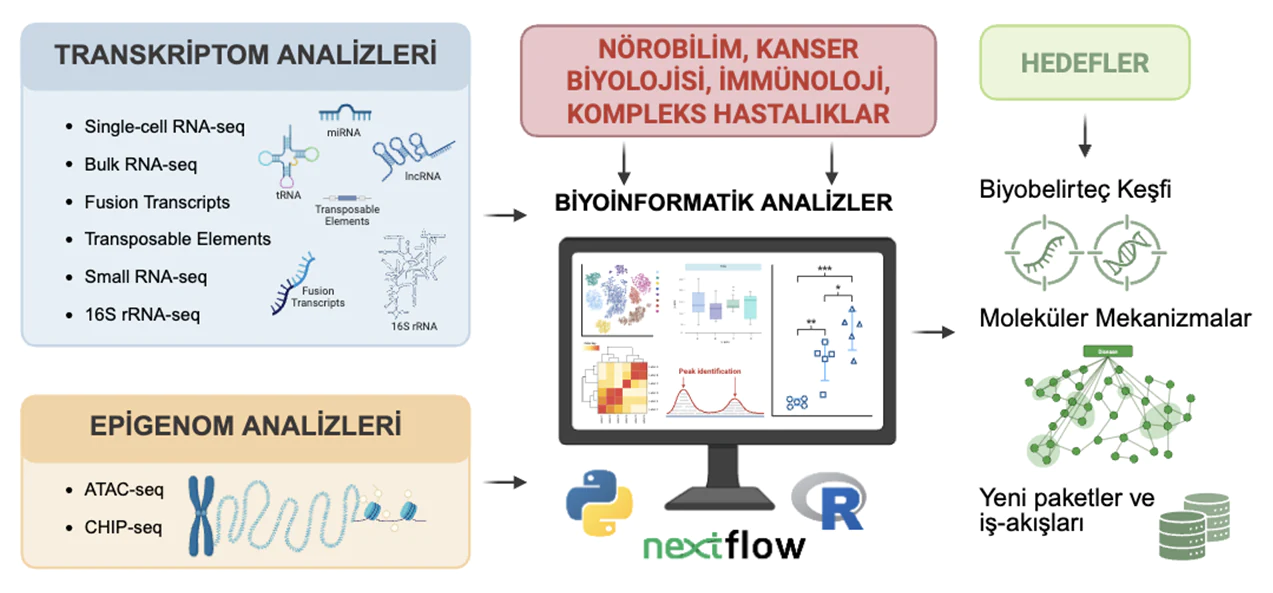
The Computational Sequence Analysis Laboratory conducts innovative research in the field of bioinformatics, focusing on the analysis of genomic, transcriptomic, and epigenomic data. Our laboratory analyzes high-throughput data generated by next-generation sequencing (NGS) technologies, including single-cell and bulk RNA-seq, fusion transcript detection, small RNAs (miRNA, tRNA, lncRNA), transposable elements, 16S rRNA sequencing, ATAC-seq, and ChIP-seq, and develops computational methods and algorithms to extract biological insights from these data types. Our studies are oriented toward biomarker discovery, molecular mechanism elucidation, gene network analysis, and the development of new software packages and automated workflows, especially in the context of complex diseases, cancer biology, immunology, and neuroscience.
Additionally, the lab is actively engaged in multi-omics data integration, data visualization, and machine learning-based modeling. Through multidisciplinary collaborations, we aim to develop novel bioinformatics solutions that advance our understanding of biological systems and support translational research.
Research Areas
- Bioinformatics
- Genomics
- Transcriptomics
- Epigenomics
- Metagenomics
Publications
Islakoğlu, Y. Ö., Korhan, P., Binokay, L., Keleş, B., Bağırsakçı, E., UludağTaşçıoğlu, M., Şamdancı, E., Karakülah, G., & Atabey, N. (2025). Fusion transcripts landscape in hepatocellular carcinoma and potential impact on the expression of fusion partners. RNA Biology, 22(1), 1–16. https://doi.org/10.1080/15476286.2025.2529036
Karacicek, B., Katkat, E., Binokay, L., Ozhan, G., Karakülah, G., &Genc, S. (2025). The role of tRNA fragments on neurogenesis alteration by H₂O₂-induced oxidative stress. Journal of Molecular Neuroscience, 75(2), 1–12. https://doi.org/10.1007/s12031-025-02299-4
Arioz, B. I., Binokay, L., Tastan, B., Genc, B., Cotuk, A., Dursun, E., Gezen-Ak, D., Hanagası, H., Gurvit, I. H., Bilgic, B., Bagriyanik, A., Karakülah, G., Yener, G. G., &Genc, S. (2025). Characterization of tRNA-derived fragments in the small neuron-derived extracellular vesicles of Alzheimer’s disease patients. Brain Research, 149730. https://doi.org/10.1016/j.brainres.2025.149730
Dayanç, B., Eris, S., Gulfirat, N. E., Ozden-Yilmaz, G., Cakiroglu, E., Coskun Deniz, O. S., Karakülah, G., Erkek-Ozhan, S., &Senturk, S. (2025). Integrative multi-omics identifies AP-1 transcription factor as a targetable mediator of acquired osimertinib resistance in non-small cell lung cancer. Cell Death & Disease, 16(1), 414. https://doi.org/10.1038/s41419-025-06888-5
Cakiroglu, E., Eris, S., Oz, O., Karakülah, G., &Senturk, S. (2025). Genome-wide CRISPR screen identifies BUB1 kinase as a druggable vulnerability in malignant pleural mesothelioma. Cell Death & Disease, 16(1), 241. https://doi.org/10.1038/s41419-025-06701-3
Projects
Ongoing Projects
TÜBİTAK - EU COST
Identification of MECOM as a Biomarker in Pancreatic Cancer and Determination of Epigenetic Inhibitor Response in MECOM-Expressing Pancreatic Cancer Cells
(Principal Investigators: S. Erkek Özhan – G. Karakülah; Researchers: Ö. Sağol, A. Aysal Ağalar, C. Ağalar, İ. T. Ünek, et al.)
TÜBİTAK 1001
The Role of the C730027H18Rik Gene in Mesenchymal-Epithelial Transition: A Novel Perspective on MET Regulation
(Principal Investigator: H. Alotaibi; Researcher: G. Karakülah) – 2024–2027
TÜSEB B Group R&D
Investigation of the Effects of Mutations in the FYR Module of the KMT2D Gene Associated with Kabuki Syndrome on Protein Interactions and Chromatin Organization
(Principal Investigator: S. Erkek Özhan; Researchers: Y. Oktay, E. Karaca Erek, G. Karakülah, A. S. Hız, G. Özden Yılmaz, et al.) – 2023–2025
Staff
Active Members
Hüseyin Güner (PhD Student)
Necati Kaan Kutlu (PhD Student)