Disease Epigenetics
Principal Investigator: Assoc. Prof. Serap ERKEK
Epigenetic dysregulation underlies a wide spectrum of disorders, from common cancers to rare genetic diseases. Our laboratory deciphers gene‑regulatory mechanisms directly at the chromatin level by integrating multi‑omics profiling with detailed biochemical and functional assays. We are especially interested in pathogenic mutations in genes encoding subunits of the MLL/COMPASS histone‑methyltransferase complexes and on how these lesions rewire regulatory networks. By characterizing the epigenetic changes associated with these mutations and identifying actionable molecular vulnerabilities, we aim to stratify patients more precisely and contribute to the development of mechanism-based therapies.
Research Areas
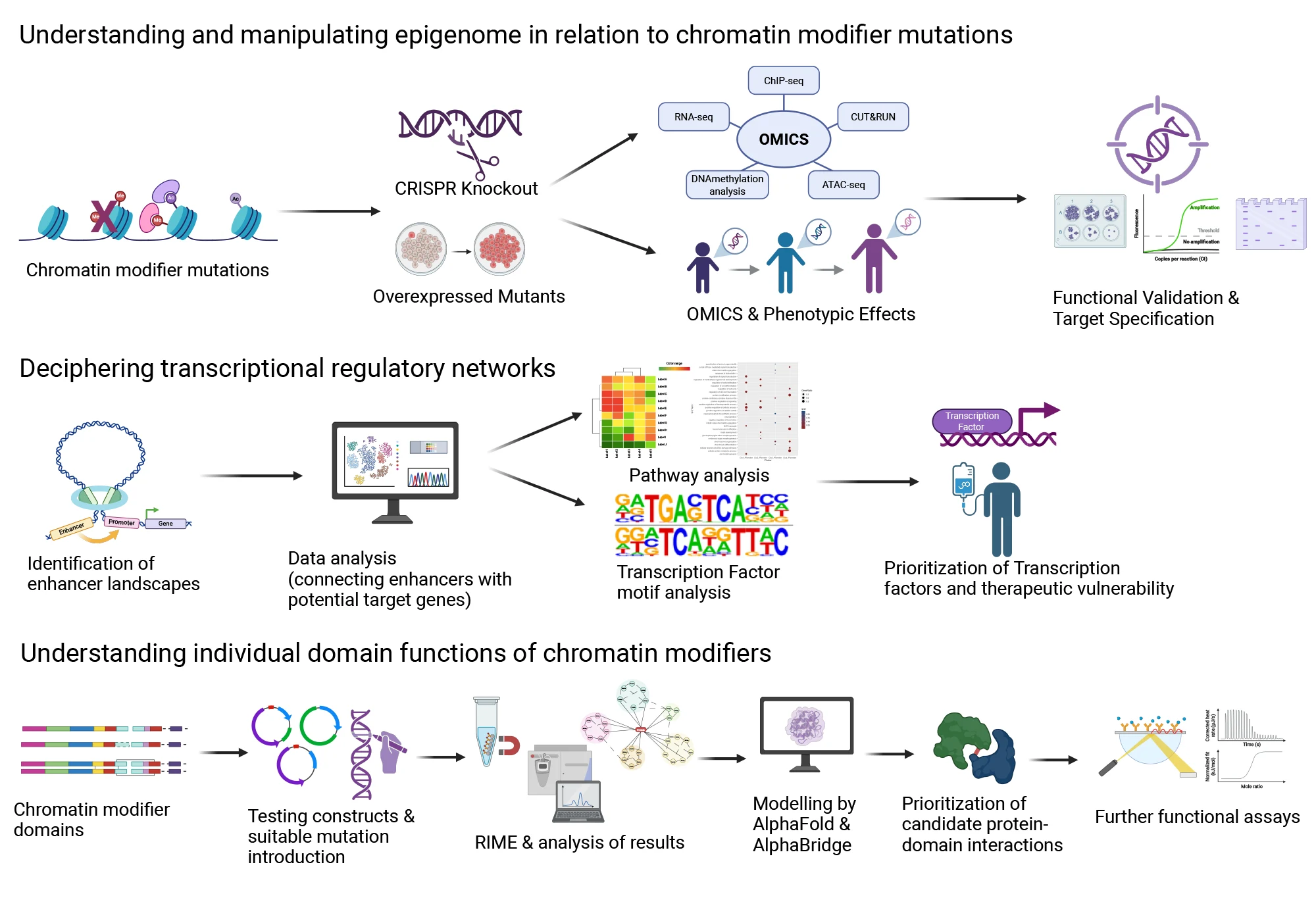
1. Understanding and manipulating epigenome in relation to chromatin modifier mutations
Multi-omics profiling (ChIP-seq, CUT&RUN-seq, RNA-seq, DNAmetylation-seq), functional assays
2. Deciphering transcriptional regulatory networks
ATAC-seq ve H3K27ac ChIP-seq, transcription factor mofif analysis, transcription factor prioritization
3. Understanding individual domain functions of chromatin modifiers
Creation of constructs expressing chromatin modifier domains, Rapid Immunoprecipitation Mass spectrometry of Endogenous proteins (RIME), identification of protein-protein interactions, prioritization using AlphaFold.
Publications
Dayanc B.* , Eris S.*, Gulfirat N.E., Ozden-Yilmaz G., Cakiroglu E., Coskun Deniz O.S., Karakülah G., Erkek-Ozhan S., Senturk S. Integrative multi-omics identifies AP-1 transcription factor as a targetable mediator of acquired osimertinib resistance in non-small cell lung cancer. Cell Death Dis. 16(1):414 (2025). *Equal first author contribution.
Akman B., Bursalı A., Gürses M., Suner A., Karakülah G., Mungan U., Yörükoğlu K., Erkek-Ozhan S. Nucleocytoplasmic β-catenin expression contributes to neuroendocrine differentiation in muscle invasive bladder cancer. Cancer Sci. 115(9):2985-2997 (2024).
Ozgun G., Yaras T., Akman B., Özden-Yılmaz G., Landman N., Karakülah G., van Lohuizen M., Senturk S., Erkek-Ozhan, S. Retinoids and EZH2 inhibitors cooperate to orchestrate cytotoxic effects on bladder cancer cells. Cancer Gene Therapy. 31(4):537-551 (2024).
Ozden-Yılmaz G., Savas B., Bursalı A., Eray A., Arıbaş A., Senturk S., Karaca E., Karakülah G., Erkek-Ozhan, S. Differential Occupancy and Regulatory Interactions of KDM6A in Bladder Cell Lines. Cells. 12(6):836 (2023).
Guneri-Sozeri, P.Y*., Ozden-Yilmaz G.*, Kisim A., Cakiroglu E., Eray A., Uzuner H., Karakülah G., Pesen Okvur D., Senturk S., Erkek-Ozhan, S. FLI1 and FRA1 transcription factors drive the transcriptional regulatory networks characterizing muscle invasive bladder cancer. Commun Biol. 6(1):199 (2023). *Equal first author contribution.
Projects
TÜBİTAK 2519 COST Action Support Program, Determination of the role of FLI1 transcription factor in tumor immunity and immunotherapy responses in bladder cancer, Ongoing
TÜBİTAK 2519 COST Action Support Program, Identification of MECOM as a biomarker in pancreatic cancer and determination of epigenetic inhibitor response in MECOM-expressing pancreatic cancer cells, Ongoing
TÜBİTAK 1001, Association of enzymatic domain-dependent functions of KDM6A protein with NF-κB signaling pathway and macrophage function in bladder cancer, Ongoing
EMBO Installation Grant, Identification of epigenetic mechanisms causing bladder cancer, Ongoing
TÜSEB Group B, Investigation of the effects of mutations occurring in the FYR module of the KMT2D gene associated with Kabuki Syndrome on protein interactions and chromatin organization, Ongoing
TÜBİTAK 2247-A National Leading Researchers Program, The role of β-catenin in neuroendocrine and neuroendocrine-like bladder tumors, Completed
TÜBİTAK 3501, Identification of transcriptional regulatory networks characterizing invasive and non-invasive bladder cancer cell lines, Completed
Staff
Active Members
Ezgi Boyvatlı
Ahmet Can Tülü
Burcu Akman
Former Members
Gizem Özgün (Doctoral Student)
Ezgi Boyvatlı (Master's Student)
Perihan Yağmur Güneri (Master's Student)
Aleyna Eray (Master's Student)